# 1 - Introduction ----------------------------------------------------
# 1.3 - Loading the tidyverse
# 1.4 - The gapminder dataset
# 2 - select() to pick columns (variables) ----------------------------
# 3 - rename() to change column names ---------------------------------
# 4 - The pipe |> -----------------------------------------------------
# 5 - filter() to pick rows (observations) ----------------------------
# Challenge 1
# Use a "pipeline" like above to output a data frame with
# columns `year`, `country`, and `lifeExp`, only for countries in Africa.
# How many rows does your output data frame have?
# 6 - arrange() to sort data frames -----------------------------------
# 7 - mutate() to modify columns and create new ones ------------------
# Challenge 2
# Use mutate() to create a new column called gdp_billion that has the
# absolute GDP (i.e., not relative to population size) and does so in
# units of billions (i.e., 2.3 would mean 2.3 billion).
# 8 - summarize() to compute summary stats ----------------------------
# Challenge 3
# Calculate the average life expectancy per country.
# Which has the longest average life expectancy and which has the
# shortest average life expectancy?Data wrangling with dplyr
1 Introduction
1.1 The dplyr package and the tidyverse
The dplyr package provides very useful functions for manipulating data in data frames (“data wrangling”). This type of data processing is often essential before you can move on to say, data visualization (next session of this workshop) or statistical data analysis.
In this session, we’ll cover the most commonly used dplyr functions:
select()to pick columns (variables)filter()to pick rows (observations)rename()to change column namesarrange()to change the order of rows (i.e., to sort a data frame)mutate()to modify values in columns and create new columnssummarize()to compute across-row summaries
All these functions take a data frame as the input, and output a new, modified data frame.
dplyr belongs to a family of R packages designed for “dataframe-centric” data science called the “tidyverse”. Dataframe-centric refers to doing most if not all data analysis while keeping the data in R’s data frame data structure. Another tidyverse package we’ll cover in today’s workshop is ggplot2 for making plots.
1.2 Setting up
Use a new script for this session – much like in the previous session:
Open a new R script (Click the
+symbol in toolbar at the top, then clickR Script)1.Save the script straight away as
data-wrangling.R– you can save it anywhere you like, though it is probably best to save it in a folder specifically for this workshop.If you want the section headers as comments in your script, like in the script I am showing you in the live session, then copy-and-paste the following into your script:
Section headers for your script (Click to expand)
1.3 Loading the tidyverse
All core tidyverse packages can be installed and loaded with a single command. Since you should already have installed the tidyverse2, you only need to load it, which you do as follows:
library(tidyverse)── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.2 ✔ tibble 3.3.0
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.1.0
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsThe output tells you which packages have been loaded as part of the tidyverse. (For now, you don’t have to worry about the “Conflicts” section.)
1.4 The gapminder dataset
In this session and the next one on data visualization, we will work with the gapminder dataset, which contains statistics such as population size for different countries across five-year intervals.
This dataset is available in a package of the same name, which you also should have already installed3. To load the package into your current R session:
# (Unlike with the tidyverse, no output is expected when you load gapminder)
library(gapminder)Take a look at the gapminder data frame that is now available to you:
gapminder# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsYou can also use the View() function to look at the data frame. This will open a new tab in your editor pane with a spreadsheet-like look and feel:
View(gapminder)
# (Should display the dataset in an editor pane tab)The gapminder data frame is a so-called “tibble”, which is the tidyverse variant of a data frame. The main difference is the nicer default printing behavior of tibbles: e.g. the data types of columns are shown, and only a limited number of rows are printed.
Each row of this data frame contains statistics for a single country in a specific year (across five-year intervals between 1952 and 2007), with the following columns:
country(stored as afactor, an alternative tocharacterthat can be used for categorical data)continent(stored as afactor)year(stored as aninteger)lifeExp(stored as aninteger): the life expectancy in yearspop(stored as aninteger): the population sizegdpPercap(stored as adouble): the per-capita GDP
2 select() to pick columns (variables)
The first dplyr function you’ll learn about is select(), which subsets a data frame by including/excluding certain columns. By default, it only includes the columns you specify:
select(.data = gapminder, year, country, pop)# A tibble: 1,704 × 3
year country pop
<int> <fct> <int>
1 1952 Afghanistan 8425333
2 1957 Afghanistan 9240934
3 1962 Afghanistan 10267083
4 1967 Afghanistan 11537966
5 1972 Afghanistan 13079460
6 1977 Afghanistan 14880372
7 1982 Afghanistan 12881816
8 1987 Afghanistan 13867957
9 1992 Afghanistan 16317921
10 1997 Afghanistan 22227415
# ℹ 1,694 more rowsAbove, the first argument was the data frame, whereas the other arguments were the (unquoted!) names of columns to be included in the output data frame4.
The order of the columns in the output data frame is exactly as you list them in select(), and doesn’t need to be the same as in the input data frame. In other words, select() is also one way to reorder columns: in the example above, we made year appear before country.
You can also specify columns that should be excluded, by prefacing their name with a ! (or a -):
# This will include all columns _except_ continent:
select(.data = gapminder, !continent)# A tibble: 1,704 × 5
country year lifeExp pop gdpPercap
<fct> <int> <dbl> <int> <dbl>
1 Afghanistan 1952 28.8 8425333 779.
2 Afghanistan 1957 30.3 9240934 821.
3 Afghanistan 1962 32.0 10267083 853.
4 Afghanistan 1967 34.0 11537966 836.
5 Afghanistan 1972 36.1 13079460 740.
6 Afghanistan 1977 38.4 14880372 786.
7 Afghanistan 1982 39.9 12881816 978.
8 Afghanistan 1987 40.8 13867957 852.
9 Afghanistan 1992 41.7 16317921 649.
10 Afghanistan 1997 41.8 22227415 635.
# ℹ 1,694 more rows3 rename() to change column names
The next dplyr function is one of the simplest: rename() to change column names.
The syntax to specify the new and old name within the function is new_name = old_name. For example, to rename the gdpPercap column:
rename(.data = gapminder, gdp_per_capita = gdpPercap)# A tibble: 1,704 × 6
country continent year lifeExp pop gdp_per_capita
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rows4 The pipe (|>)
Examples so far applied a single dplyr function to a data frame, simply printing the output (a new data frame) to screen. But in practice, it’s common to use several consecutive dplyr functions to wrangle a dataframe into the format you want.
For example, you may want to first select() one or more columns, and then rename() a column. You could do that as follows:
gapminder_sel <- select(.data = gapminder, year, country, gdpPercap)
rename(.data = gapminder_sel, gdp_per_capita = gdpPercap)# A tibble: 1,704 × 3
year country gdp_per_capita
<int> <fct> <dbl>
1 1952 Afghanistan 779.
2 1957 Afghanistan 821.
3 1962 Afghanistan 853.
4 1967 Afghanistan 836.
5 1972 Afghanistan 740.
6 1977 Afghanistan 786.
7 1982 Afghanistan 978.
8 1987 Afghanistan 852.
9 1992 Afghanistan 649.
10 1997 Afghanistan 635.
# ℹ 1,694 more rowsAnd you could go on along these lines, successively creating new objects that you then use for the next step.
But there is a more elegent way of dong this, directly sending (“piping”) output from one function into the next function with the pipe operator |> (a vertical bar | followed by a greater-than sign >). Let’s start by seeing a reformulation of the code above with pipes:
gapminder |>
select(year, country, gdpPercap) |>
rename(gdp_per_capita = gdpPercap)# A tibble: 1,704 × 3
year country gdp_per_capita
<int> <fct> <dbl>
1 1952 Afghanistan 779.
2 1957 Afghanistan 821.
3 1962 Afghanistan 853.
4 1967 Afghanistan 836.
5 1972 Afghanistan 740.
6 1977 Afghanistan 786.
7 1982 Afghanistan 978.
8 1987 Afghanistan 852.
9 1992 Afghanistan 649.
10 1997 Afghanistan 635.
# ℹ 1,694 more rowsWhat happened here? We took the gapminder data frame, sent (“piped”) it into the select() function, whose output in turn was piped into the rename() function. You can think of the pipe as “then”: take gapminder, then select, then rename.
When using the pipe, you no longer specify the input data frame with the .data argument, because the function now gets its input data via the pipe (specifically, the input goes to the function’s first argument by default).
5 filter() to pick rows (observations)
5.1 Introduction
The filter() function outputs only those rows that satisfy one or more conditions. It is similar to Filter functionality in Excel — except that those only change what you display, while filter() will completely exclude rows.
But if that sounds scary, recall that dplyr functions always output a new data frame. Therefore, with any dplyr function, you’ll only modify existing data when you assign the output back to the input object, like in this example with the select() function:
# [Don't run this - hypothetical example]
# After running this, any columns in the 'cats' dataframe other than 'name'
# and 'coat' would be removed from it:
cats <- cats |> select(name, coat)5.2 Filter based on one condition
This first filter() example outputs only rows for which the life expectancy exceeds 80 years (remember, each row represents a country in a given year):
gapminder |>
filter(lifeExp > 80)# A tibble: 21 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Australia Oceania 2002 80.4 19546792 30688.
2 Australia Oceania 2007 81.2 20434176 34435.
3 Canada Americas 2007 80.7 33390141 36319.
4 France Europe 2007 80.7 61083916 30470.
5 Hong Kong, China Asia 2002 81.5 6762476 30209.
6 Hong Kong, China Asia 2007 82.2 6980412 39725.
7 Iceland Europe 2002 80.5 288030 31163.
8 Iceland Europe 2007 81.8 301931 36181.
9 Israel Asia 2007 80.7 6426679 25523.
10 Italy Europe 2002 80.2 57926999 27968.
# ℹ 11 more rowsHow many rows were output? (Click to see the answer)
21 rows were output, as shown in the first line above: 21 x 6 (rows x columns)
As the example above demonstrated, filter() outputs rows that satisfy the condition(s) you specify. These conditions don’t have to be based on numeric comparisons:
# Only keep rows where the value in the 'continent' column is 'Europe':
gapminder |>
filter(continent == "Europe")# A tibble: 360 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Albania Europe 1952 55.2 1282697 1601.
2 Albania Europe 1957 59.3 1476505 1942.
3 Albania Europe 1962 64.8 1728137 2313.
4 Albania Europe 1967 66.2 1984060 2760.
5 Albania Europe 1972 67.7 2263554 3313.
6 Albania Europe 1977 68.9 2509048 3533.
7 Albania Europe 1982 70.4 2780097 3631.
8 Albania Europe 1987 72 3075321 3739.
9 Albania Europe 1992 71.6 3326498 2497.
10 Albania Europe 1997 73.0 3428038 3193.
# ℹ 350 more rows== to test for equality!
5.3 Filter based on multiple conditions
It’s also possible to filter based on multiple conditions. For example, you may want to see which countries in Asia had a life expectancy greater than 80 years in 2007:
gapminder |>
filter(continent == "Asia", year == 2007, lifeExp > 80)# A tibble: 3 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Hong Kong, China Asia 2007 82.2 6980412 39725.
2 Israel Asia 2007 80.7 6426679 25523.
3 Japan Asia 2007 82.6 127467972 31656.Like above, by default, multiple conditions are combined using a Boolean AND. In other words, in a given row, each condition must be met to output the row.
If you want to combine conditions using a Boolean OR, where only one of the conditions needs to be met, use a | between the conditions:
# Keep rows with a high life expectancy and/or a high GDP:
gapminder |>
filter(lifeExp > 80 | gdpPercap > 10000)# A tibble: 392 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Argentina Americas 1977 68.5 26983828 10079.
2 Argentina Americas 1997 73.3 36203463 10967.
3 Argentina Americas 2007 75.3 40301927 12779.
4 Australia Oceania 1952 69.1 8691212 10040.
5 Australia Oceania 1957 70.3 9712569 10950.
6 Australia Oceania 1962 70.9 10794968 12217.
7 Australia Oceania 1967 71.1 11872264 14526.
8 Australia Oceania 1972 71.9 13177000 16789.
9 Australia Oceania 1977 73.5 14074100 18334.
10 Australia Oceania 1982 74.7 15184200 19477.
# ℹ 382 more rows5.4 Pipeline practice
Finally, let’s practice another time with “pipelines” that use multiple dplyr functions: filter rows, then select columns, and finally rename one of the remaining columns:
gapminder |>
filter(continent == "Americas") |>
select(year, country, gdpPercap) |>
rename(gdp_per_capita = gdpPercap)# A tibble: 300 × 3
year country gdp_per_capita
<int> <fct> <dbl>
1 1952 Argentina 5911.
2 1957 Argentina 6857.
3 1962 Argentina 7133.
4 1967 Argentina 8053.
5 1972 Argentina 9443.
6 1977 Argentina 10079.
7 1982 Argentina 8998.
8 1987 Argentina 9140.
9 1992 Argentina 9308.
10 1997 Argentina 10967.
# ℹ 290 more rowsChallenge 1
Use a “pipeline” like above to output a data frame with columns year, country, and lifeExp, only for countries in Africa. How many rows does your output data frame have?
Click for the solution
gapminder |>
filter(continent == "Africa") |>
select(year, country, lifeExp)# A tibble: 624 × 3
year country lifeExp
<int> <fct> <dbl>
1 1952 Algeria 43.1
2 1957 Algeria 45.7
3 1962 Algeria 48.3
4 1967 Algeria 51.4
5 1972 Algeria 54.5
6 1977 Algeria 58.0
7 1982 Algeria 61.4
8 1987 Algeria 65.8
9 1992 Algeria 67.7
10 1997 Algeria 69.2
# ℹ 614 more rows6 arrange() to sort data frames
The arrange() function is like sorting functionality in Excel: it changes the order of rows based on the values in one or more columns6.
gapminder is currently first sorted alphabetically by country, and next by year. But you may, for example, want to sort by population size instead:
gapminder |>
arrange(pop)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Sao Tome and Principe Africa 1952 46.5 60011 880.
2 Sao Tome and Principe Africa 1957 48.9 61325 861.
3 Djibouti Africa 1952 34.8 63149 2670.
4 Sao Tome and Principe Africa 1962 51.9 65345 1072.
5 Sao Tome and Principe Africa 1967 54.4 70787 1385.
6 Djibouti Africa 1957 37.3 71851 2865.
7 Sao Tome and Principe Africa 1972 56.5 76595 1533.
8 Sao Tome and Principe Africa 1977 58.6 86796 1738.
9 Djibouti Africa 1962 39.7 89898 3021.
10 Sao Tome and Principe Africa 1982 60.4 98593 1890.
# ℹ 1,694 more rowsSorting can help you find observations with the smallest or largest values for a certain column: above, we see that the smallest population size in the dataset is Sao Tome and Principe in 1952.
Default sorting is from small to large, as seen above . To sort in reverse order, use the desc() (descending) helper function:
gapminder |>
arrange(desc(pop))# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 China Asia 2007 73.0 1318683096 4959.
2 China Asia 2002 72.0 1280400000 3119.
3 China Asia 1997 70.4 1230075000 2289.
4 China Asia 1992 68.7 1164970000 1656.
5 India Asia 2007 64.7 1110396331 2452.
6 China Asia 1987 67.3 1084035000 1379.
7 India Asia 2002 62.9 1034172547 1747.
8 China Asia 1982 65.5 1000281000 962.
9 India Asia 1997 61.8 959000000 1459.
10 China Asia 1977 64.0 943455000 741.
# ℹ 1,694 more rowsFinally, you may want to sort by multiple columns, where ties in the first column are broken by a second column (and so on). Do this by simply listing the columns in the appropriate order:
# Sort first by continent, then by country:
gapminder |>
arrange(continent, country)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Algeria Africa 1952 43.1 9279525 2449.
2 Algeria Africa 1957 45.7 10270856 3014.
3 Algeria Africa 1962 48.3 11000948 2551.
4 Algeria Africa 1967 51.4 12760499 3247.
5 Algeria Africa 1972 54.5 14760787 4183.
6 Algeria Africa 1977 58.0 17152804 4910.
7 Algeria Africa 1982 61.4 20033753 5745.
8 Algeria Africa 1987 65.8 23254956 5681.
9 Algeria Africa 1992 67.7 26298373 5023.
10 Algeria Africa 1997 69.2 29072015 4797.
# ℹ 1,694 more rows7 mutate() to modify columns and create new ones
So far, we’ve focused on functions that subset and reorganize data frames (and you’ve seen how to modify column names). But you haven’t seen how you can change the data or compute derived data.
This can be done with the mutate() function. For example, to create a new column that has population sizes in millions rather than in individuals:
# Create a new column 'pop_million' by dividing 'pop' by a million:
gapminder |>
mutate(pop_million = pop / 10^6)# A tibble: 1,704 × 7
country continent year lifeExp pop gdpPercap pop_million
<fct> <fct> <int> <dbl> <int> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779. 8.43
2 Afghanistan Asia 1957 30.3 9240934 821. 9.24
3 Afghanistan Asia 1962 32.0 10267083 853. 10.3
4 Afghanistan Asia 1967 34.0 11537966 836. 11.5
5 Afghanistan Asia 1972 36.1 13079460 740. 13.1
6 Afghanistan Asia 1977 38.4 14880372 786. 14.9
7 Afghanistan Asia 1982 39.9 12881816 978. 12.9
8 Afghanistan Asia 1987 40.8 13867957 852. 13.9
9 Afghanistan Asia 1992 41.7 16317921 649. 16.3
10 Afghanistan Asia 1997 41.8 22227415 635. 22.2
# ℹ 1,694 more rowsTo modify an existing column rather than adding a new one, simply “assign back to the same name”:
# Change the unit of the 'pop' column:
gapminder |>
mutate(pop = pop / 10^6)# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8.43 779.
2 Afghanistan Asia 1957 30.3 9.24 821.
3 Afghanistan Asia 1962 32.0 10.3 853.
4 Afghanistan Asia 1967 34.0 11.5 836.
5 Afghanistan Asia 1972 36.1 13.1 740.
6 Afghanistan Asia 1977 38.4 14.9 786.
7 Afghanistan Asia 1982 39.9 12.9 978.
8 Afghanistan Asia 1987 40.8 13.9 852.
9 Afghanistan Asia 1992 41.7 16.3 649.
10 Afghanistan Asia 1997 41.8 22.2 635.
# ℹ 1,694 more rowsChallenge 2
Use mutate() to create a new column gdp_billion that has the absolute GDP (i.e., not relative to population size) in units of billions (i.e., 2.3 would mean 2.3 billion).
Click for the solution
You need to do two things, which can be combined into a single line:
- Make the GDP absolute by multiplying by the population size:
gdpPercap * pop - Change the unit of the absolute GDP to billions by dividing by a billion:
/ 10^9
gapminder |>
mutate(gdp_billion = gdpPercap * pop / 10^9)# A tibble: 1,704 × 7
country continent year lifeExp pop gdpPercap gdp_billion
<fct> <fct> <int> <dbl> <int> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779. 6.57
2 Afghanistan Asia 1957 30.3 9240934 821. 7.59
3 Afghanistan Asia 1962 32.0 10267083 853. 8.76
4 Afghanistan Asia 1967 34.0 11537966 836. 9.65
5 Afghanistan Asia 1972 36.1 13079460 740. 9.68
6 Afghanistan Asia 1977 38.4 14880372 786. 11.7
7 Afghanistan Asia 1982 39.9 12881816 978. 12.6
8 Afghanistan Asia 1987 40.8 13867957 852. 11.8
9 Afghanistan Asia 1992 41.7 16317921 649. 10.6
10 Afghanistan Asia 1997 41.8 22227415 635. 14.1
# ℹ 1,694 more rows8 summarize() to compute summary stats
The final function dplyr function we’ll cover is summarize(), which computes summaries of your data across rows. For example, to calculate the mean GDP across the entire dataset (all rows):
# The syntax is similar to 'mutate': <new-column> = <operation>
gapminder |>
summarize(mean_gdp = mean(gdpPercap))# A tibble: 1 × 1
mean_gdp
<dbl>
1 7215.The output is still a dataframe, but unlike with all previous dplyr functions, it is completely different from the input dataframe, “collapsing” the data down to as little as a single number, like above.
summarize() becomes really powerful in combination with the helper function group_by() to compute groupwise stats. For example, to get the mean GDP separately for each continent:
gapminder |>
group_by(continent) |>
summarize(mean_gdp = mean(gdpPercap))# A tibble: 5 × 2
continent mean_gdp
<fct> <dbl>
1 Africa 2194.
2 Americas 7136.
3 Asia 7902.
4 Europe 14469.
5 Oceania 18622.group_by() implicitly splits a data frame into groups of rows: here, one group for observations from each continent. After that, operations like in summarize() will happen separately for each group, which is how we ended up with per-continent means.
Challenge 3
Calculate the average life expectancy per country. Which country has the longest average life expectancy and which has the shortest average life expectancy?
Click for some hints
Since you’ll need to examine your output in two ways, it makes sense to store the output of the life expectancy calculation in a dataframe.
Next, an easy way to get rows with lowest and highest values in a column is to simply sort by that column and examining the output.
Click for the solution
First, create a dataframe with the mean life expectancy by country:
lifeExp_bycountry <- gapminder |>
group_by(country) |>
summarize(mean_lifeExp = mean(lifeExp))Then, arrange that dataframe in two directions to see the countries with the longest and shortest life expectancy. You could optionally pipe into head() to only see the top n, here top 1:
lifeExp_bycountry |>
arrange(mean_lifeExp) |>
head(n = 1)# A tibble: 1 × 2
country mean_lifeExp
<fct> <dbl>
1 Sierra Leone 36.8lifeExp_bycountry |>
arrange(desc(mean_lifeExp)) |>
head(n = 1)# A tibble: 1 × 2
country mean_lifeExp
<fct> <dbl>
1 Iceland 76.59 Bonus material for self-study
9.1 Writing and reading tabular data to and from files
When working with your own data in R, you’ll usually need to read data from files into your R environment, and conversely, to write data that is in your R environment to files.
While it’s possible to make R interact with Excel spreadsheet files7, storing your data in plain-text files generally benefits reproducibility. Tabular plain text files can be stored using:
- A Tab as the column delimiter (often called TSV files, and stored with a
.tsvextension) - A comma as the column delimiter (often called CSV files, and stored with a
.csvextension).
To practice writing and reading data to and from TSV files, the examples below use functions from the readr package. This package is part of the core tidyverse and therefore already loaded into your environment.
Writing to files
To write the gapminder data frame to a TSV file, use the write_tsv() function with argument x for the R object and argument file for the file path:
write_tsv(x = gapminder, file = "gapminder.tsv")Because we simply provided a file name, the file will have been written in your R working directory8.
If you want to take a look at the file you just created, you can find it in RStudio’s Files tab and click on it, which will open it in the editor panel. Of course, you could also find it in your computer’s file browser and open it in different ways.
The file argument to write_tsv() takes a file path, meaning that you can specify any location on your computer for it, in addition to the file name.
For example, if you had a folder called results in your current working directory, you could save the file in there like so:
write_tsv(x = gapminder, file = "results/gapminder.tsv")Note that a forward slash / as a folder delimiter will work regardless of your operating system (even though Windows natively delimits folder by a backslash \).
Finally, if you want to store the file in a totally different place than your working dir, it’s good to know that you can also use a so-called absolute (or “full”) path. For example:
write_tsv(x = gapminder, file = "/Users/poelstra.1/Desktop/gapminder.tsv")Reading from files
To practice reading data from a file, use the read_tsv() function on the file you just created:
gapminder_reread <- read_tsv(file = "gapminder.tsv")Rows: 1704 Columns: 6
── Column specification ────────────────────────────────────────────────────────
Delimiter: "\t"
chr (2): country, continent
dbl (4): year, lifeExp, pop, gdpPercap
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.Note that this function is rather chatty, telling you how many rows and columns it read, and what their data types are. Let’s check the resulting object:
gapminder_reread# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rowsThis looks good! Do note that the column’s data types are not identical to what they were (year and pop are saved as double rather than integer, and country and continent as character rather than factor). This is largely expected because that kind of metadata is not stored in a plain-text TSV, so read_tsv() will by default simply make best guesses as to the types.
Alternatively, you could tell read_tsv() what the column types should be, using the col_types argument (with abbreviations f for factor, i for integer, d for double — run ?read_tsv for more info):
read_tsv(file = "gapminder.tsv", col_types = "ffidi")# A tibble: 1,704 × 6
country continent year lifeExp pop gdpPercap
<fct> <fct> <int> <dbl> <int> <dbl>
1 Afghanistan Asia 1952 28.8 8425333 779.
2 Afghanistan Asia 1957 30.3 9240934 821.
3 Afghanistan Asia 1962 32.0 10267083 853.
4 Afghanistan Asia 1967 34.0 11537966 836.
5 Afghanistan Asia 1972 36.1 13079460 740.
6 Afghanistan Asia 1977 38.4 14880372 786.
7 Afghanistan Asia 1982 39.9 12881816 978.
8 Afghanistan Asia 1987 40.8 13867957 852.
9 Afghanistan Asia 1992 41.7 16317921 649.
10 Afghanistan Asia 1997 41.8 22227415 635.
# ℹ 1,694 more rows9.2 Counting rows with count() and n()
A common operation is simply counting the number of observations for each group. The dplyr package comes with two related functions that help with this.
For instance, to check the number of countries included in the dataset for the year 2002, you can use the count() function. It takes the name of one or more columns to define the groups we are interested in, and can optionally sort the results in descending order with sort = TRUE:
gapminder |>
filter(year == 2002) |>
count(continent, sort = TRUE)# A tibble: 5 × 2
continent n
<fct> <int>
1 Africa 52
2 Asia 33
3 Europe 30
4 Americas 25
5 Oceania 2If you need to use the number of observations in calculations, the n() function is useful. It will return the total number of observations in the “current group” as applicable – for instance, to get the standard error of life expectancy per continent:
gapminder |>
group_by(continent) |>
summarize(se_life = sd(lifeExp) / sqrt(n()))# A tibble: 5 × 2
continent se_life
<fct> <dbl>
1 Africa 0.366
2 Americas 0.540
3 Asia 0.596
4 Europe 0.286
5 Oceania 0.7759.3 Function name conflicts
When you loaded the tidyverse, the output included a “Conflicts” section that may have seemed ominous:
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsWhat this means is that two tidyverse functions, filter() and lag(), have the same names as two functions from the stats package that were already in your R environment.
Those stats package functions are part of what is often referred to as “base R”: core R functionality that is always available (loaded) when you start R.
Due to this function name conflict/collision, the filter() function from dplyr “masks” the filter() function from stats. Therefore, if you write a command with filter(), it will use the dplyr function and not the stats function.
You can use a “masked” function, by prefacing it with its package name as follows: stats::filter().
9.4 Using the pipe keyboard shortcut
The following RStudio keyboard shortcut will insert a pipe symbol: Ctrl/Cmd + Shift + M.
However, by default this (still) inserts an older pipe symbol, %>%. As long as you’ve loaded the tidyverse9, it would not really be a problem to just use %>% instead of |>. But it would be preferred to make RStudio insert the |> pipe symbol:
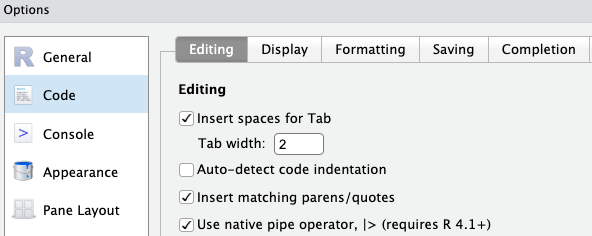
- Click
Tools>Global Options>Codetab on the right - Check the box “Use native pipe operator” as shown below
- Click
OKat the bottom of the options dialog box to apply the change and close the box.
9.5 Learn more
The material in this page was adapted from this Carpentries lesson episode, which has a lot more content than we were able to cover today.
Regarding data wrangling specifically, here are two particularly useful additional skills to learn:
Pivoting/reshaping — moving between ‘wide’ and ‘long’ data formats with
pivot_wider()andpivot_longer(). This is covered in episode 13 of the focal Carpentries lesson.Joining/merging — combining multiple dataframes based on one or more shared columns. This can be done with dplyr’s
join_*()functions – see for example this chapter in R for Data Science.
Footnotes
Or Click
File=>New file=>R Script.↩︎If not: run
install.packages("tidyverse")now.↩︎If not: run
install.packages("gapminder")now.↩︎Based on what you learned in the previous session, it may seem strange that the column names should not be quoted.
One way to make sense of this is that the columns can be thought of as (vector) objects, whose names are the column names.↩︎Using pipes is also faster and uses less computer memory.↩︎
It will always rearrange the order of rows as a whole, never just of individual columns since that would scramble the data.↩︎
Using the readxl package. This is installed as part of the tidyverse but is not a core tidyverse package and therefore needs to be loaded separately:
library(readxl).↩︎Recall that you can see where that is at the top of the Console tab, or by running
getwd().↩︎This symbol only works when you have one of the tidyverse packages loaded.↩︎